MAR Contextual databases
Data collection
The MarRef, MarDb, MarFun, and SalDB contextual databases are built by aggregating data from a number of public available sequence, taxonomy and literature databases in a semi-automatic fashion. Other databases or resources such as bacterial diversity and culture collections databases, web mapping service and ontology databases are used extensively for curation of metadata as shown in the figure below.
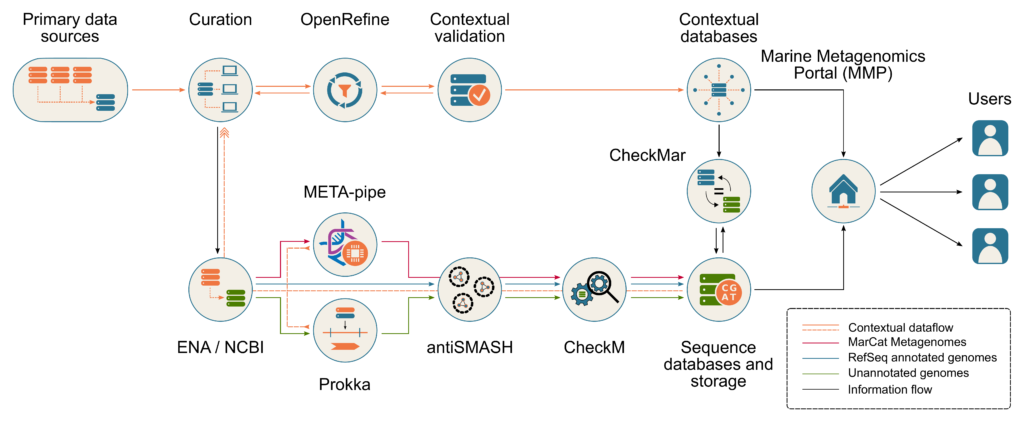
The contextual MAR databases, supports the International community-driven standards of the Genomics Standards Consortium and is fully compliant with its recommendations for Minimum Information about any (x) Sequence (MIxS) standards, including MIGS (Minimum Information about a Genome Sequence), MISAG (Minimum information about a single amplified genome) and MIMAG (a metagenome-assembled genome) of bacteria and archaea and MIMS (Minimal Information of Metagenome sequence). The databases have also included the proposed standards for provenance of analysis developed in the ELIXIR-EXCELERATE project (The metagenomic data life-cycle: standards and best practices).
Our databases contain each more than 150 metadata attributes providing information about sampling environment, the organism and taxonomy, phenotype, pathogenicity, secondary metabolites, assembly and annotation. When applicable, attributes makes use of well established ontologies to keep free text to a strict minimum, and links to external resources whenever possible.
Curation, refinement and validation
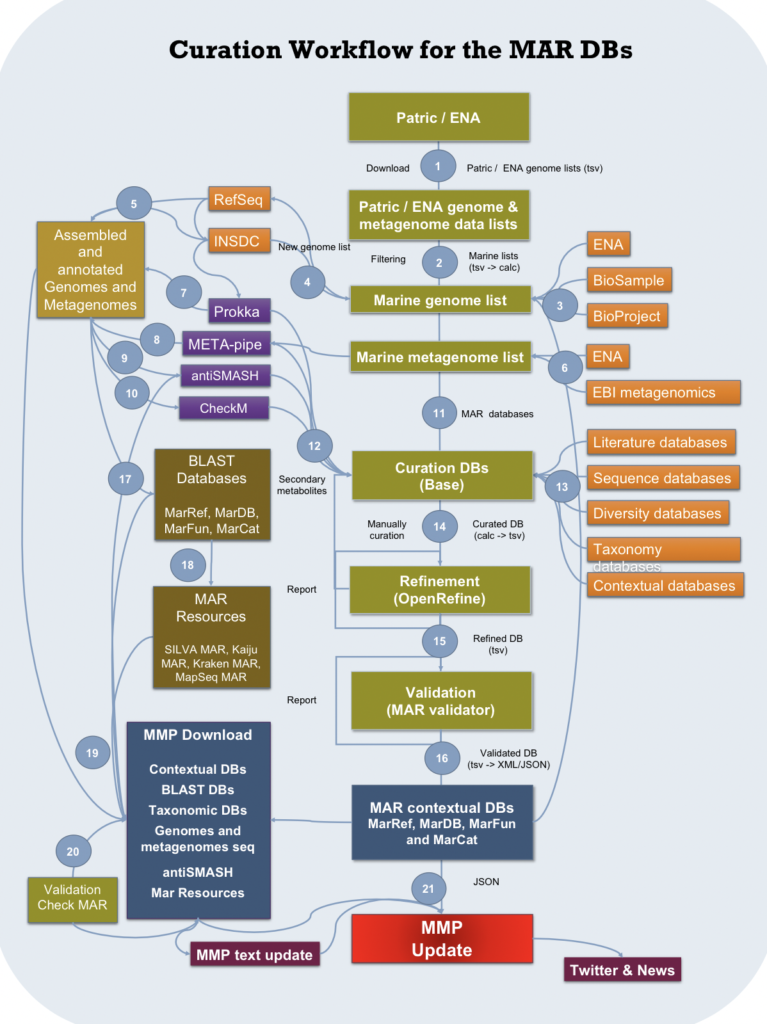
The curation process is essential to ensure that all manually annotated entries are handled in a consistent manner. Each record undergoes a critical review of experimental and predicted data as well as manual verification of information from the literature. The records are continuously updated as new information becomes available.
OpenRefine is used for refining the metadata attributes by cleaning, trimming of leading and trailing whitespace, transforming data from one format into another and extending it with web services and external data. Before being stored in a MongoDB database, the curated data is validated using a JSON schema to ensure consistency and help curators catching overlooked errors. The whole curation process is shown in the figure below.
The MAR Sequence Databases
MarRef, MarDB and SalDB sequence databases
MarRef and MarDB sequence databases are primarily built on gene, protein and genome sequences obtained from the Prokaryotic RefSeq Genomes database. All archaeal and bacterial genomes in RefSeq have been annotated using the NCBI’s Prokaryotic Genome Automatic Annotation Pipeline, PGAAP. However, ∼20% of all records in MarDB did not have any RefSeq entry with PGAAP annotations. Circumventing the lack of gene and protein information of these genomes, annotation was performed on pre-assembled sequences using Prokka, a command line software tool, for annotation of prokaryotic genomes.
MarFun sequence database
Databases derived from the MarRef, MarDB and SalDB
From MarRef and MarDB, several sequence databases has been generated to enhance taxonomic classification and functional assignment of metagenomics samples. SILVAmar is a subset database of the SILVA database , which provides comprehensive, quality checked of aligned small (16S SSU) ribosomal RNA (rRNA) sequences for bacteria and archaea, based on the curated entries in MarRef and MarDB. A 16S rRNA database for MAPseq, a set of fast and accurate sequence read classification tools designed to assign taxonomy and OTU classifications, based on MarRef and MarDB has also been generated. The Kaijumar protein sequence database has been implemented in Kaiju, a program for sensitive taxonomic classification of high-throughput sequencing reads from metagenomic experiments. The METAmar sequence database, was established to increase the functional assignment of protein coding sequences (CDS`s) in marine metagenomics samples, consists of all predicted genes in MarRef and MarDB and contain approx. 40 million entries.
Secondary metabolites
Secondary metabolites was predicted by using the bacterial version (v4.1.0) of antiSMASH 4. The antiSMASH results can be assessed from MarRef, MarDB and SalDB either by using the link to the antiSMASH results for each record or by searching all records in the Browse meny in MarRef or MarDB. For more info regarding the antiSMASH output please follow this link. All records has also been mapped to ChEBI and ChEMBL.
Update scheme
The MAR databases is updated bi-anually, usually in January/February and August/September.


